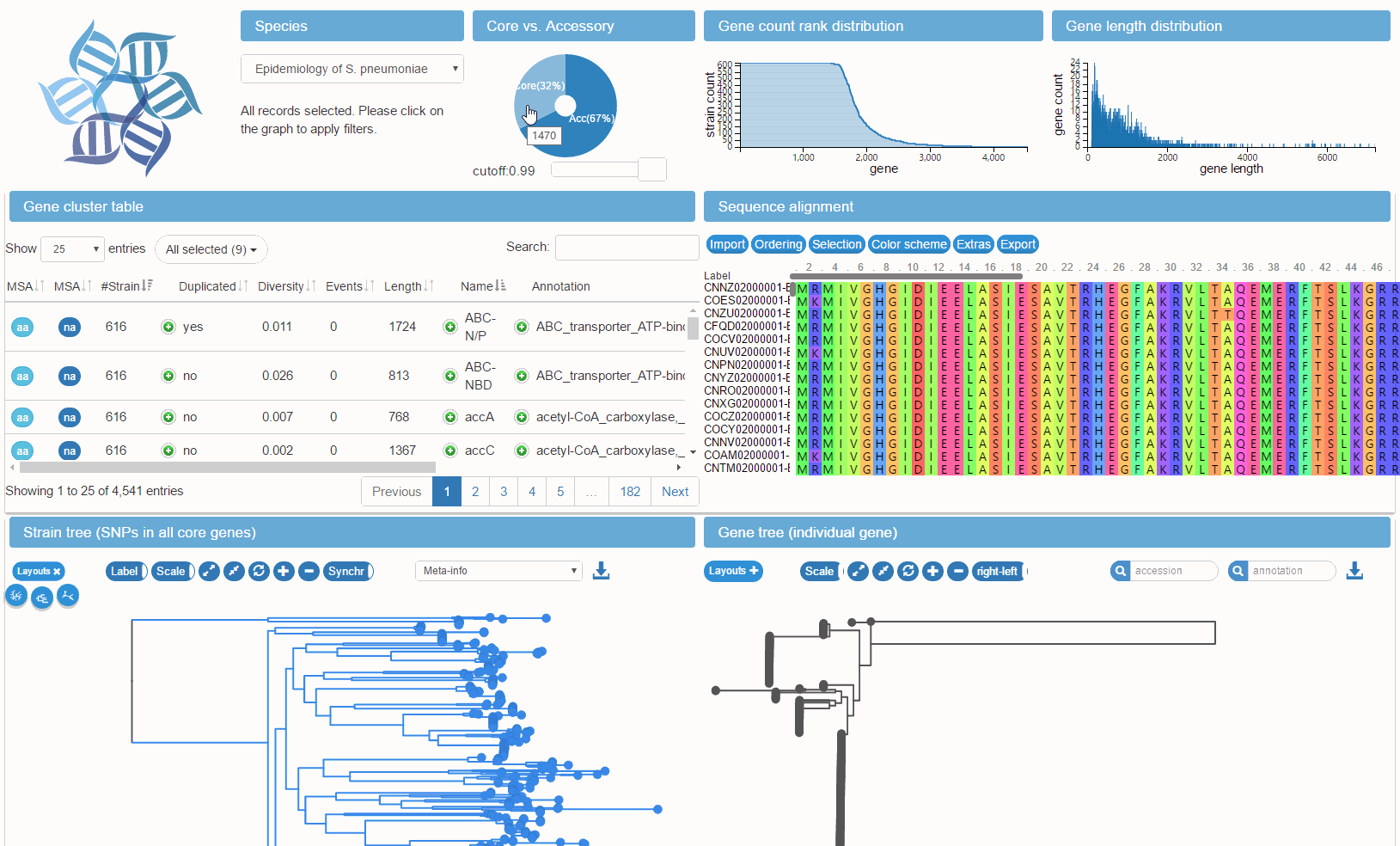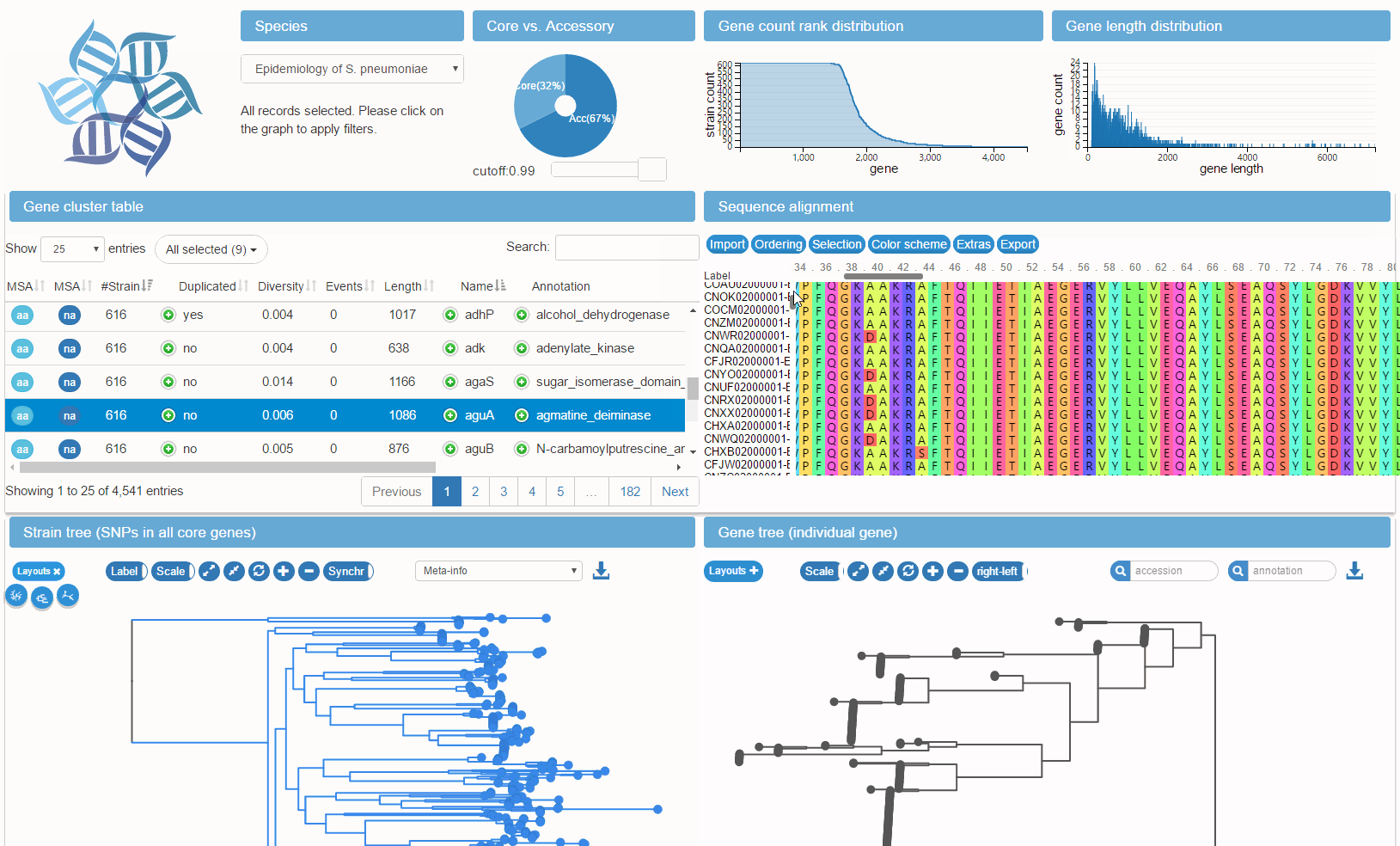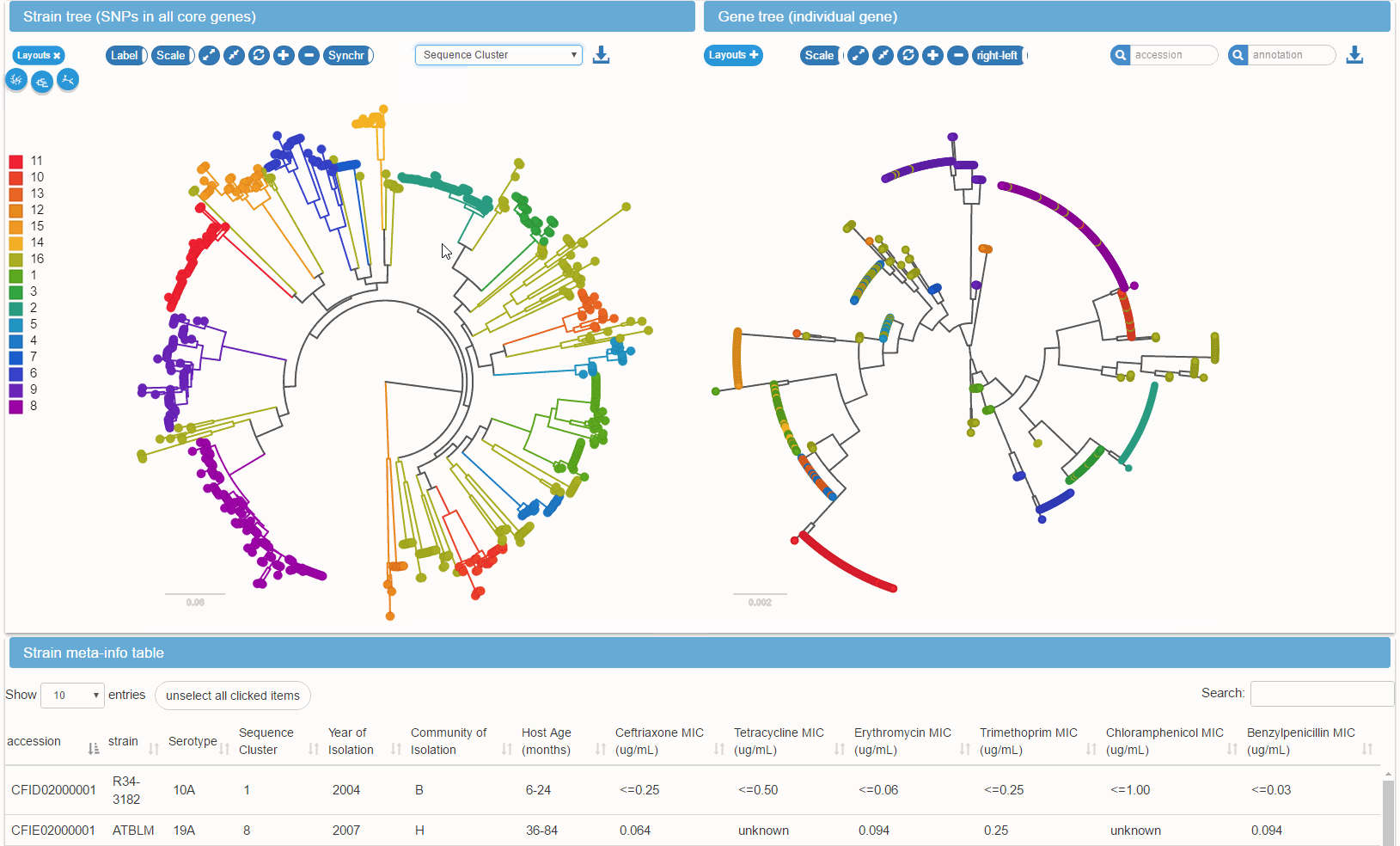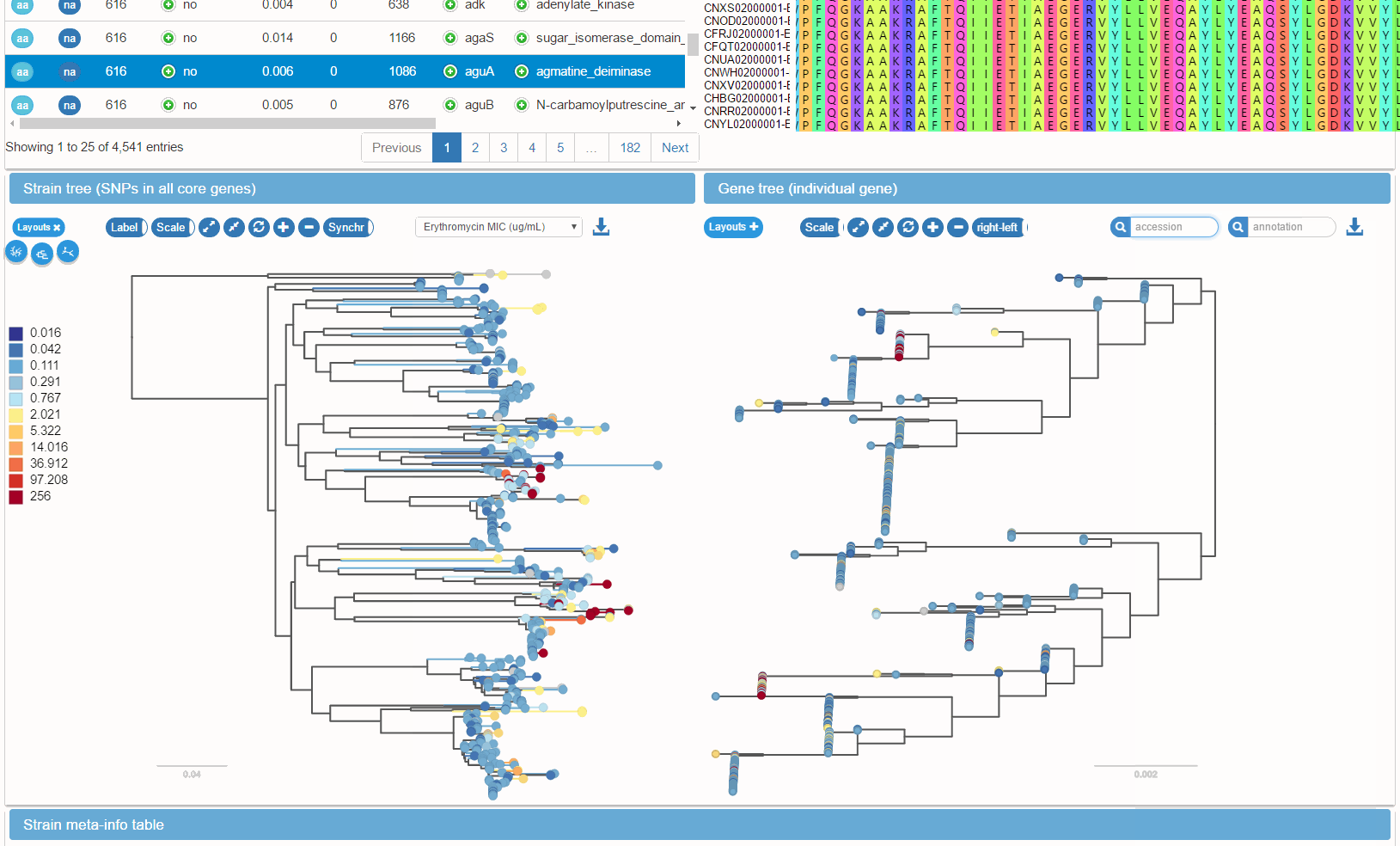
panX is a software package for comprehensive analysis, interactive visualization and dynamic exploration of bacterial pan-genomes. The analysis pipeline is based on DIAMOND, MCL and phylogeny-aware post-processing.
The visualization application encompasses various interconnected components (statistical charts, gene cluster table, alignment, comparative phylogenies, metadata). Gene clusters can be rapidly searched and filtered by summary statistics such as annotation and phylogenetic properties such as diversity.
Metadata and gene presence/absence patterns can be mapped onto the species tree. Such mapping facilitates the identification of genes associated with phenotypes such as antibiotic resistance, virulence, or epidemiological parameters such as host age.
Showcases of panX functionalities:
Explore another example: diverse Prochlorococcus genomes
If you use panX, please cite:
Wei Ding, Franz Baumdicker, Richard A Neher; panX: pan-genome analysis and exploration, Nucleic Acids Research, Volume 46, Issue 1, 9 January 2018, Pages e5,
https://doi.org/10.1093/nar/gkx977